University of New South Wales researchers have demonstrated that hydrogen can be released and reabsorbed from sodium borohydride, a promising storage material, overcoming a major hurdle to i
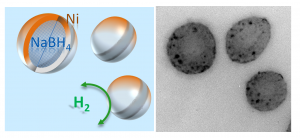
A diagram of the nanoparticle, with sodium borohydride encased in nickel, and a TEM image of the particles (credit: University of New South Wales)
ts use as an alternative fuel source.
Considered a major a fuel of the future, hydrogen could be used to power buildings, portable electronics and vehicles — but this application hinges on practical storage technology.
The researchers synthesized nanoparticles of sodium borohydride and encased these inside nickel shells.
Their unique “core-shell” nanostructure demonstrated remarkable hydrogen storage properties, including the release of energy at much lower temperatures than previously observed.
“No one has ever tried to synthesize these particles at the nanoscale because they thought it was too difficult, and couldn’t be done. We’re the first to do so, and demonstrate that energy in the form of hydrogen can be stored with sodium borohydride at practical temperatures and pressures,” says Dr Kondo-Francois Aguey-Zinsou from the School of Chemical Engineering at UNSW.
Lightweight compounds known as borohydrides (including lithium and sodium compounds) are known to be effective storage materials, but it was believed that once the energy was released it could not be reabsorbed — a critical limitation. This perceived “irreversibility” means there has been little focus on sodium borohydride.
“By controlling the size and architecture of these structures we can tune their properties and make them reversible — this means they can release and reabsorb hydrogen,” says Aguey-Zinsou. “We now have a way to tap into all these borohydride materials, which are particularly exciting for application on vehicles because of their high hydrogen storage capacity.”
In its bulk form, sodium borohydride requires temperatures above 550 degrees Celsius just to release hydrogen. However, with the core-shell nanostructure, the researchers saw initial energy release happening at just 50 °C, and significant release at 350 °C.
“The new materials that could be generated by this exciting strategy could provide practical solutions to meet many of the energy targets set by the U.S. Department of Energy,” says Aguey-Zinsou.





 sending...
sending...
